[TEMPLATE] R: Start here
This is a preview with example data.
Welcome to a Synthesize Bio notebook!
These are notebooks that allow you to create custom analysis on any datasets. You can then re-run any notebook on new data.
This tutorial teaches you about some of the magic functionality that makes our notebooks unique.
This is an R notebook. Please also try a Python notebook!
What Makes This Notebook Different?
- Special functions for easily displaying interactive plots and tables
- Automatic data import as an
SummarizedExperimentobject - Notebook sharing and re-running
Built in functions for this notebook environment
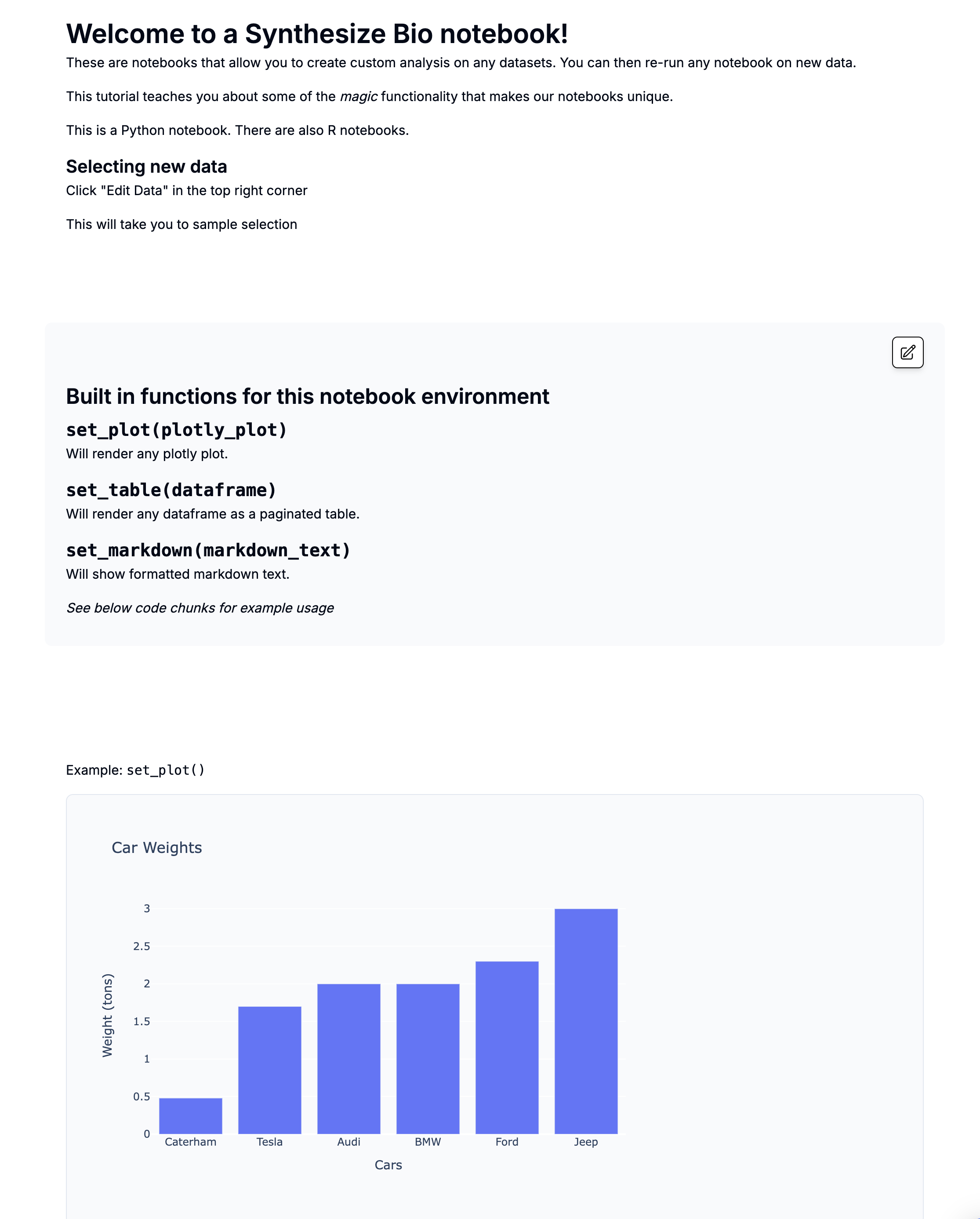
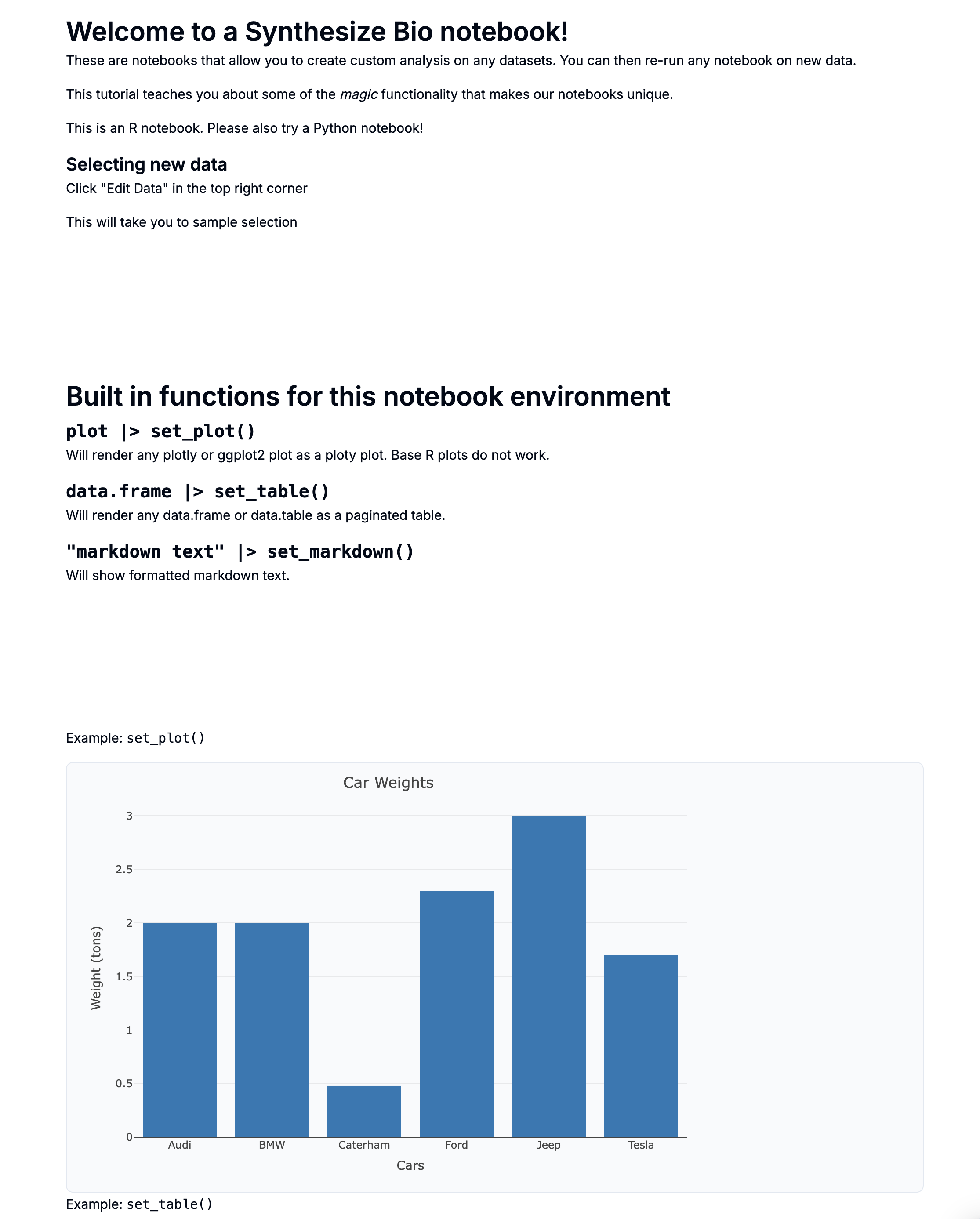
plot |> set_plot()
Will render any plotly or ggplot2 plot as a ploty plot. Base R plots do not render interactively, but can still be displayed.
data.frame |> set_table()
Will render any data.frame or data.table as a paginated table.
"markdown text" |> set_markdown()
Will show formatted markdown text.
See the code chunk below for example usage.
Data is Loaded as a SummarizedExperiment Object
See the below chunk for how the summarized experiment object is loaded
This offers several advantages:
- Sample-level metadata is available in
colData(). - Gene-level metadata (e.g., mapping identifiers) is available in
rowData(). - Raw counts are stored in
assay()and TPM inassay()$TPM.
When working with SummarizedExperiment objects, keep in mind that colData() and rowData()
objects are not data.frame objects and so they need to be converted to data.frame for set_table() to work
Editing
- You can freely modify this notebook.
- Add, edit, or rearrange code and text chunks as needed.
Editing Code Chunks
- Toggle "View code" in the header to display all code.
- Run individual chunks by clicking the ► button in the top-left corner of each chunk.
- Execute the entire notebook by clicking ► in the header.
Code Editor Shortcuts
This notebook supports VS Code-style shortcuts for efficient editing:
- Press F1 to open the command palette and view all commands.
- Comment/uncomment a code selection with Command (⌘) + /.
- Indent code with Command (⌘) + ].
- Move chunks using the Edit menu in the top-left corner of each chunk.
Sharing and Re-running
Share your notebook using the 🌍 Share button in the top-right menu to generate a read-only link for colleagues.
You can also use the top-right menu to:
- Create a copy of the notebook.
- Edit Data and re-run the notebook on different datasets.