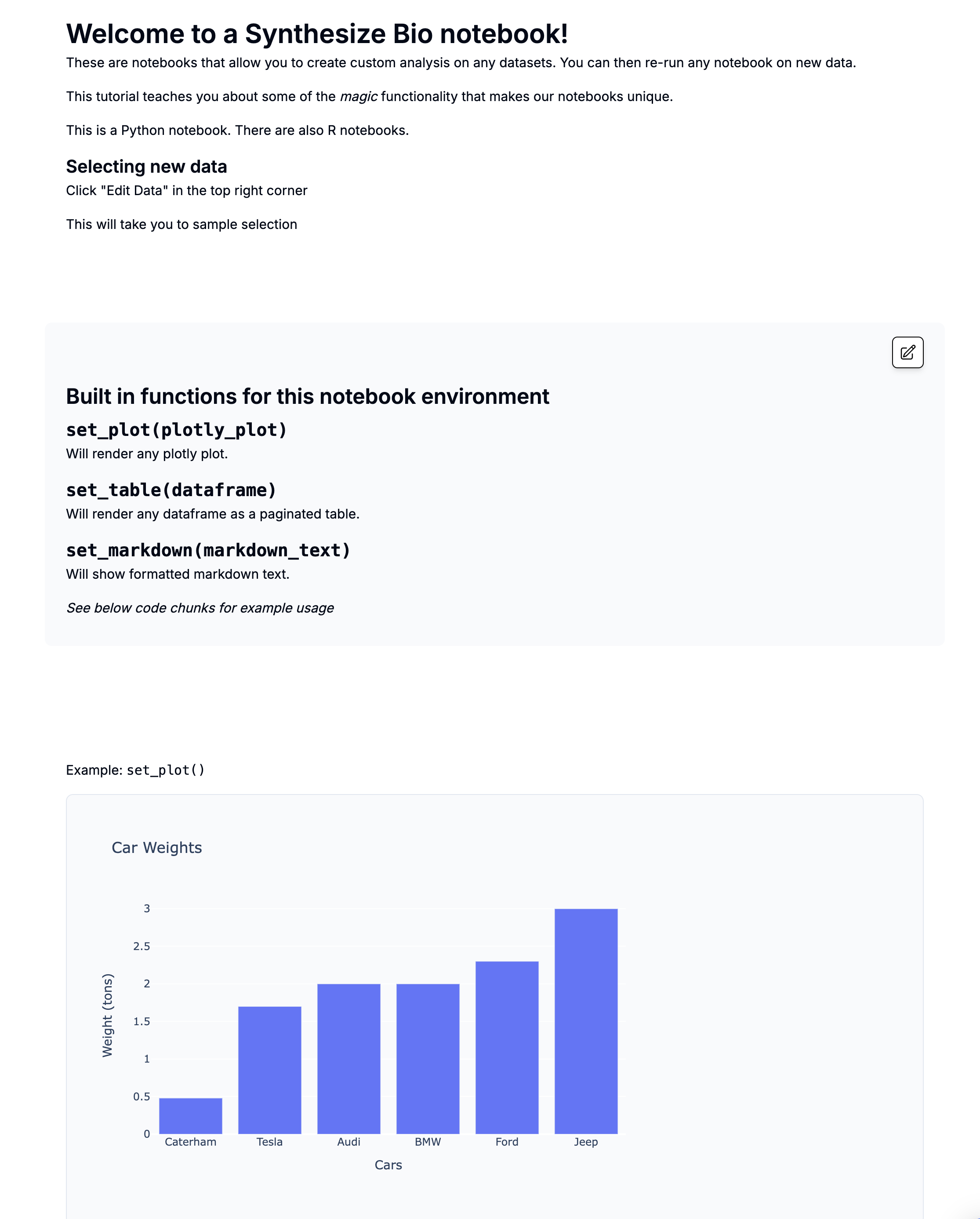
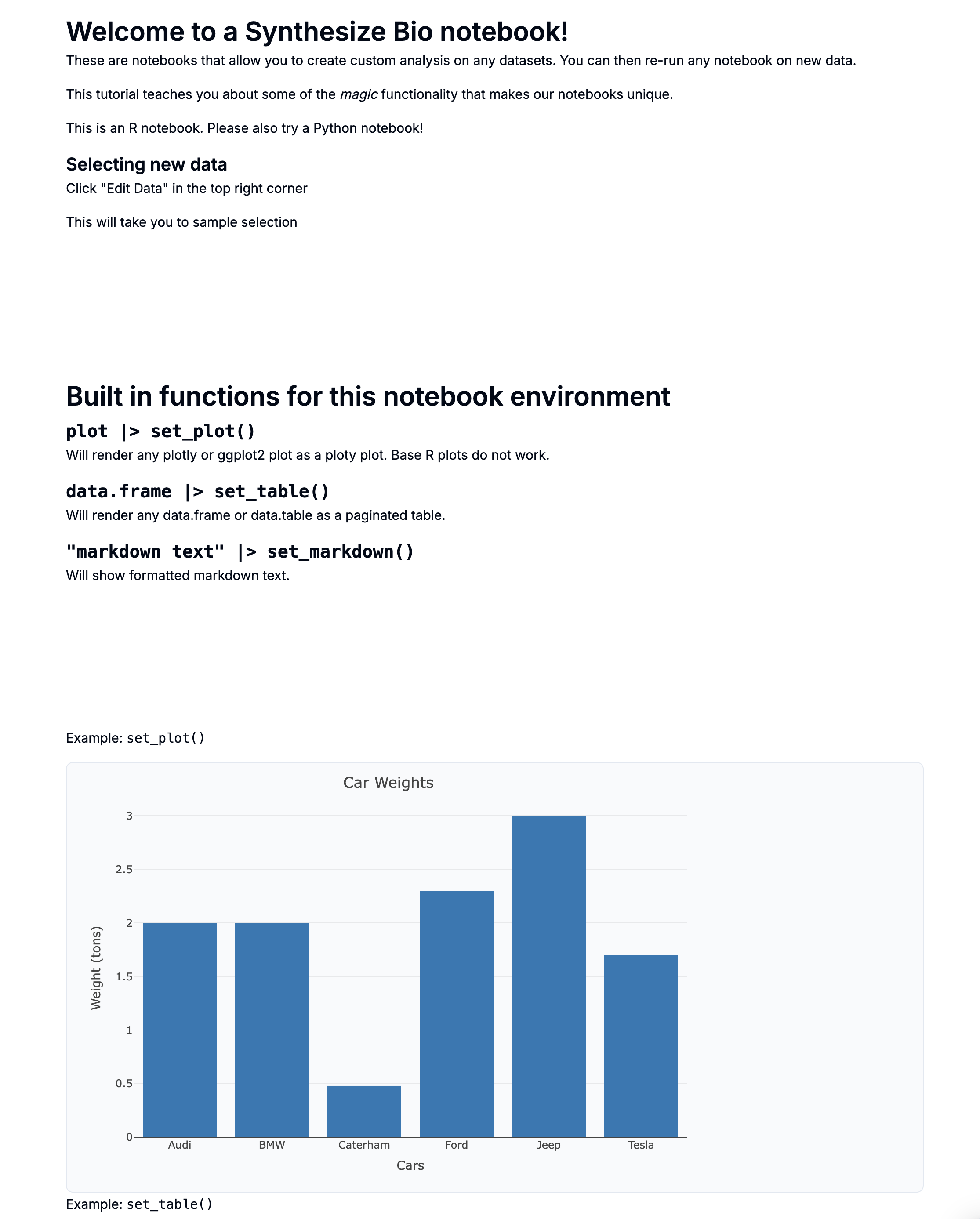
Overexpression example from demo
Overexpression example from demo
Introduction
This notebook demonstrates RNA-seq differential expression analysis following the workflow developed by Michael I. Love and colleagues, as described in their widely-used DESeq2 package and associated publications. The analysis includes gene-level exploratory data analysis and differential expression testing.
For simplicity, we've selected a subset of the analyses present in the paper. You can refer to the paper for thorough explanations of each step by clicking on the link icon (🔗) in each section header.
Synthesize.bio will build the SummarizedExperiment object for you. You can explore or generate your own data by clicking on "Edit Data" and selecting new datasets and groups of samples. This analysis requires 2 sample groups.
Load packages
Our R kernel has some pre-installed packages that we load here. Refer to our "R: Start here" template for more information about pre-installed packages.
Import Data
We can import the SummarizedExperiment object generated by the Synthesize.bio backend using our load_data API, run some checks, and display our DataFrame.
Note the SummarizedExperiment variable group_labels providing our predefined sample groups.
Construct a DESeqDataSet 🔗
We can construct a DESeqDataSet object from our SummarizedExperiment that will then form the starting point of the analysis.
Exploratory analysis and visualization 🔗
Pre-filtering the dataset
We select genes with at least 1 count in the dataset.
PCA plot 🔗
Using the top 1000 highly variable genes.
We diverge from the paper a bit here by using Variance Stabilizing Transformation instead of rlog for more than 30 samples to speed things up.
Differential expression analysis 🔗
Running the differential expression pipeline
Plotting results 🔗
Plotting normalized counts over intervention
Gene clustering 🔗
Heatmap of transformed values across samples for the top 20 highly-variable genes.
Gene Set Enrichment Analysis
There are many methods to accomplish gene set enrichment analysis. Here we use an over-representation test (a.k.a hypergeometric test) implemented with the {gprofiler2} R package.
Exporting results 🔗
You can download this csv at the footer of the table.
References
Love MI, Anders S, Kim V and Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression [version 2; peer review: 2 approved]. F1000Research 2016, 4:1070 https://doi.org/10.12688/f1000research.7035.2