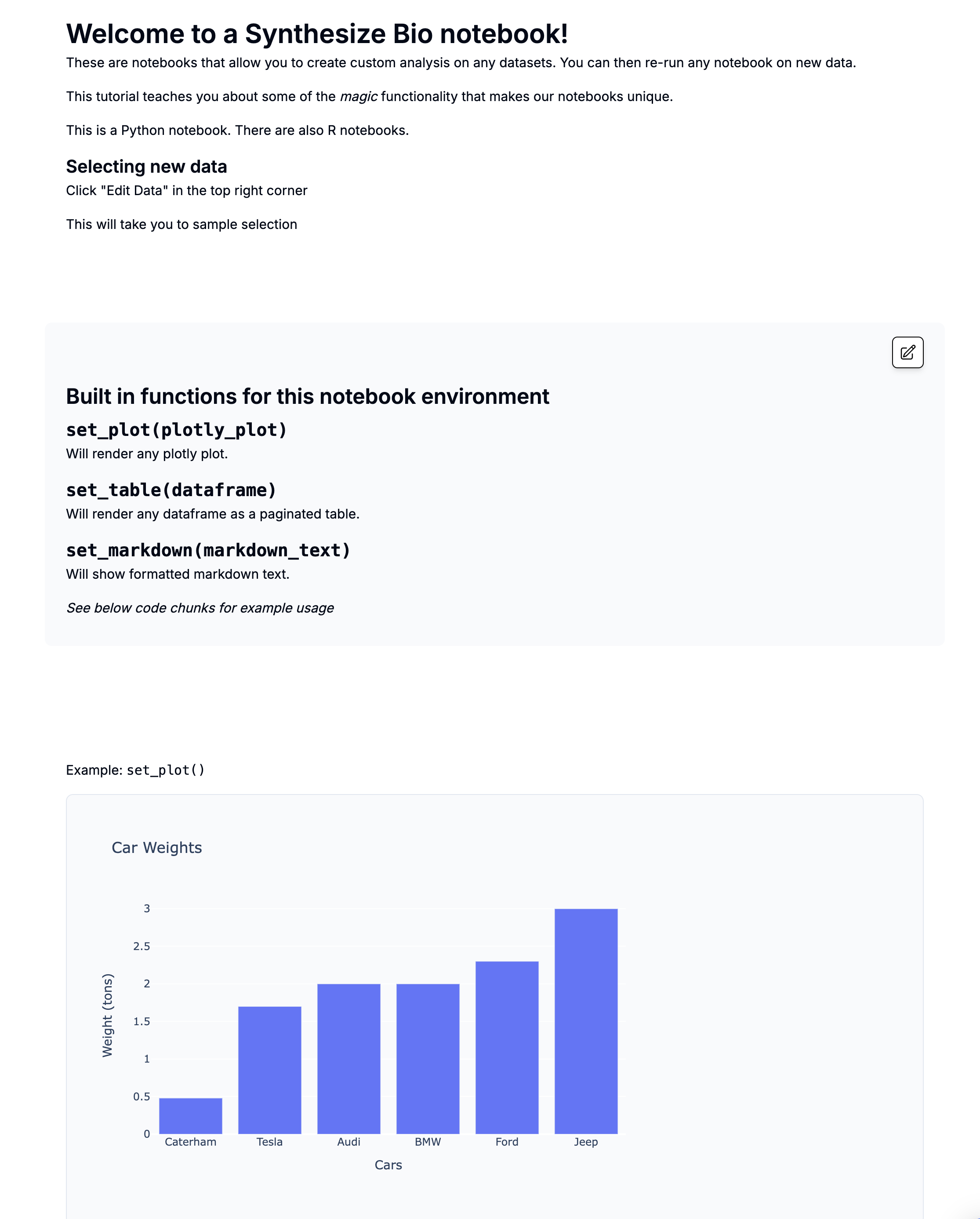
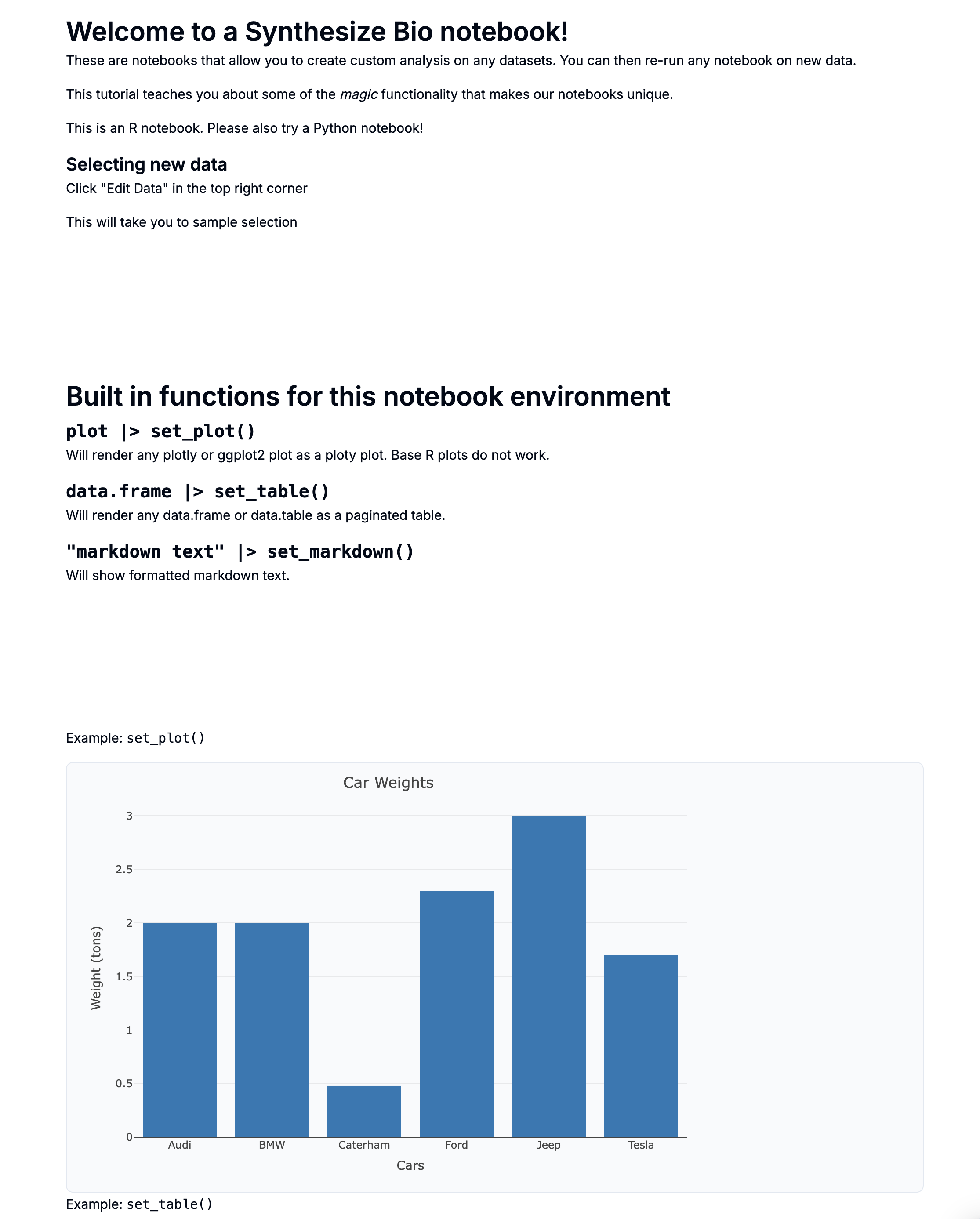
[TEMPLATE] Comparing two groups with pydeseq2
This is a preview with example data.
Introduction
This notebook performs differential expression analysis between two sample groups using DESeq2 in Python.
You can run by selecting new data
- Edit data at the top right
- This will take you to sample selection
- Please select two groups of samples - they will be directly compared!
- Note that this example compares "responders" to "non-responders" but you can change these sample group names to fit your design
Data Summary
This is a summary of the samples within our responders and non-responders sample groups.
PCA plot
Here we plot the scores for each sample for PC1 and PC2 using the top 3000 highly variable genes.
DESeq2 Results
Here is a results table showing differential expression analysis between responders and non-responders. The table includes base mean expression, log2 fold changes, and adjusted p-values for 60,684 genes.
Volcano Plot
Volcano plot showing the distribution of differentially expressed genes between responders and non-responders. Each point represents a gene, with log2 fold change on the x-axis and statistical significance (-log10 p-value) on the y-axis.
P-value distribution histogram
The p-value histogram serves as a key diagnostic visualization. For the most meaningful interpretation, it's recommended to filter out low-count genes from this plot, as these genes with minimal expression can create artificial peaks in the histogram distribution. This is why we typically only include genes with mean normalized counts above a threshold (e.g., > 1).
Centered Gene Expression Heatmap
To create a more informative visualization than raw expression values, we can plot how each gene's expression deviates from its mean across all samples by centering the data (subtracting each gene's average).
The resulting heatmap reveals sample-specific patterns of up- and down-regulation that might be obscured in absolute expression data.
Here we plot log2-transformed transcript per million (TPM) values.
Gene set enrichment results
Gene set enrichment analysis results showing significantly enriched biological pathways and processes between responders and non-responders. The analysis identifies which functional gene sets are over-represented in the differentially expressed genes.
This is the end of this analysis. You can add more code chunks here or go to the top and "Edit Data" or "copy notebook" to create a new analysis.