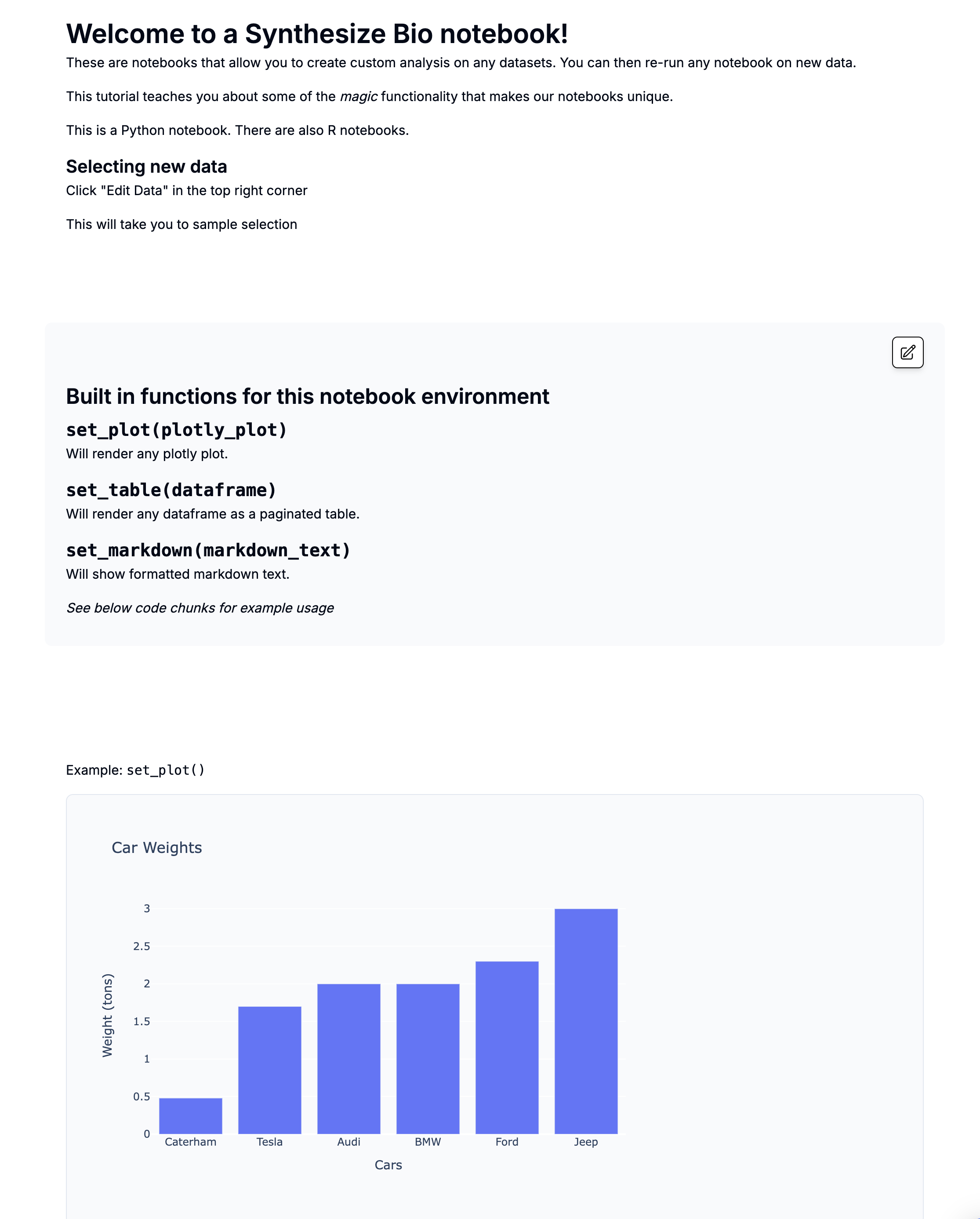
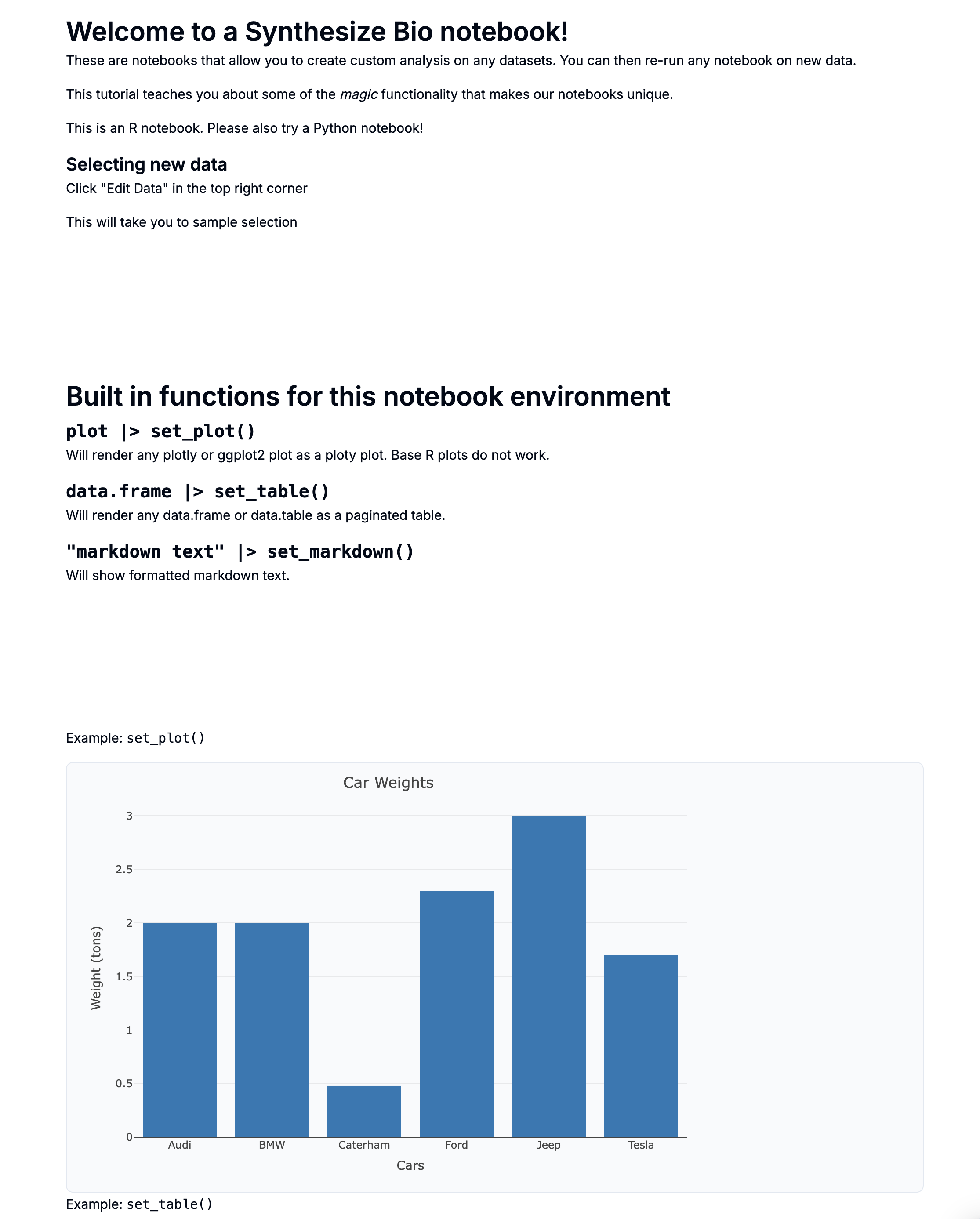
Intro to Synthesize Bio Generative Models
[TEMPLATE] Intro to Synthesize Bio Generative Models
- AI Dataset Simple Sample Generation Example
- AI Dataset Simple mean estimation example
This is a preview with example data.
What is AI-generated data?
Synthesize Bio has developed multi-modal deep learning models capable of generating gene expression data.
If you're familiar with image-generation models like Stable Diffusion, Midjourney, or DALLE, our models work in a similar way. They are trained on large datasets annotated with sample-specific information.
Just as you might ask an image-generation model to create a photo of lawn flamingos on the moon, you can prompt our models to generate RNA-seq data for specific cell lines under various perturbations or to simulate single-cell RNA-seq data from different tissues.
We are drafting formal publications describing the development and performance of our models. In the meantime, this notebook has some frequently-asked questions.
Model types
There are two model types/modes available today to test:
- Mean estimation: These models create a distribution capturing the biological heterogeneity consistent with the supplied metadata. This distribution is then sampled to predict a gene expression distribution that captures measurement error. The mean of that distribution serves as the prediction
- Sample generation: This model works identically to the mean estimation approach, except that the final gene expression distribution is also sampled to generate realistic-looking synthetic data that captures the error associated with measurements
We are actively training new models that will be released under early access plans. Our models can generate different data modalities (for example, single-cell or bulk RNA-seq).
Mean estimation models should be preferred in use cases whose primary purpose is to reliably estimate an effect of interest as they only sample biological variability.
In contrast, sample generation should be preferred when generating data with the same “look and feel” as experimental data. This can also be important if training downstream models that will be applied to real data.
In the below heatmap, each column is an AI-generated sample. Mouse over the labels to see which model type was used.